Spatial Transcriptomics to Proteomics Prediction (STP Challenge)
STP Open Challenge - Benchmarking Spatial Transriptomics-to-Proteomics Prediction
GitHub: stpoc_gnn
This project develops a multimodal deep learning pipeline for the Spatial Transcriptomics-to-Proteomics (STP) Challenge, which aims to infer spatial protein expression from H&E images and RNA-seq profiles in glioma tissue. The task requires models to integrate heterogeneous spatial signals, handle irregular tissue geometry, and generalize across patients and measurement modalities.
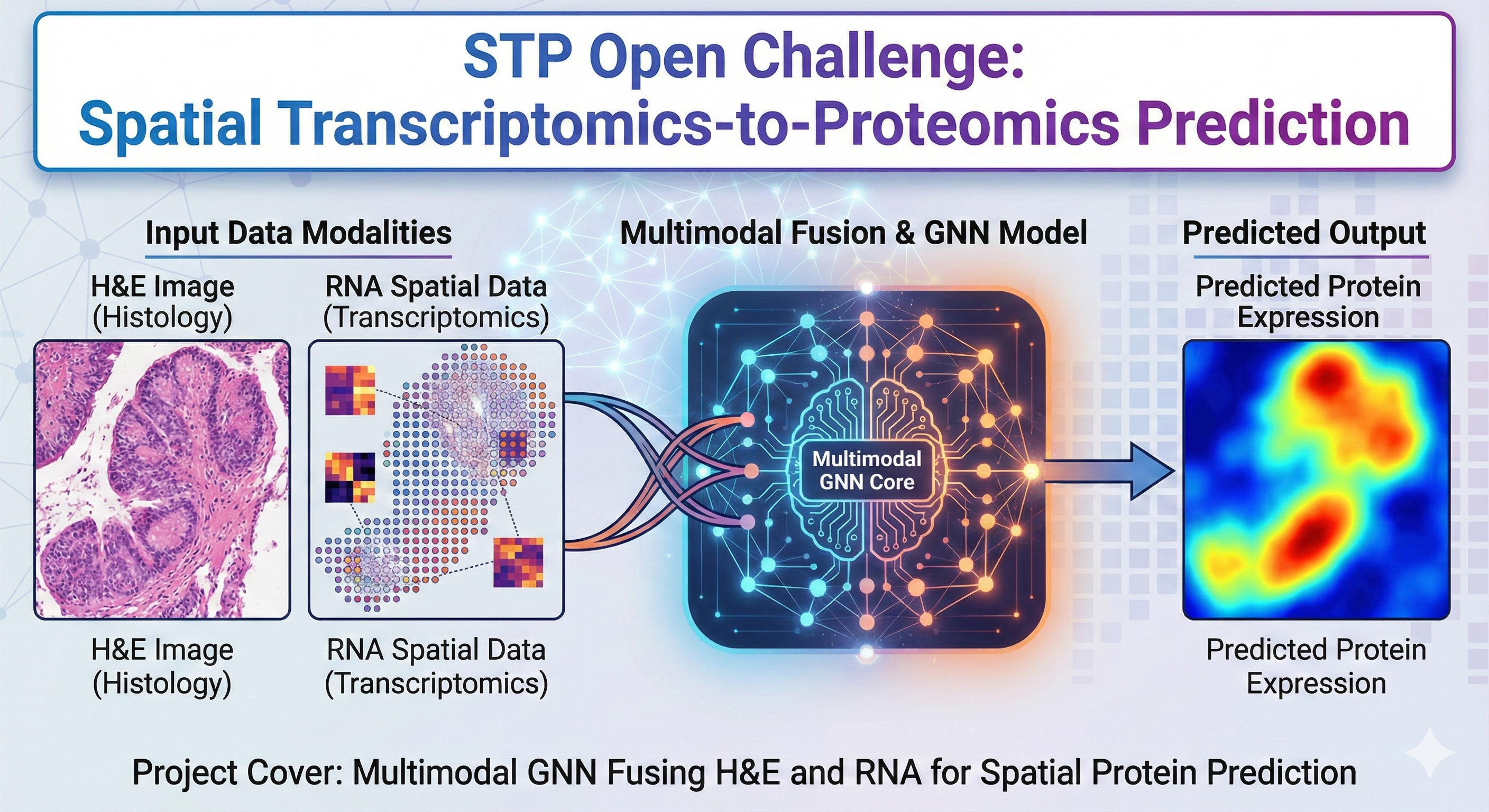
Preprocessing and Data Integration
We implemented a complete preprocessing pipeline to unify the spatial-omics inputs:
- Patch-based H&E feature extraction: Each tissue slide is partitioned into fixed-size patches aligned to Visium spot coordinates. A pretrained ResNet / ConvNeXt feature encoder transforms each patch into compact morphological embeddings.
- RNA normalization & embedding: Raw transcript counts are log-normalized, scaled, and projected through a gene embedding MLP to stabilize cross-sample variation.
- Spatial graph construction: Spots are represented as nodes in a graph where edges follow spatial proximity (kNN or radius-based). This captures tissue architecture and morphological continuity.
- Multimodal fusion: Visual and transcriptomic embeddings are concatenated or adaptively gated to support cross-modality information flow.
Modeling Approach
We designed several model architectures and performed systematic ablations:
- Baseline CNN-only model: A UNet-style image-to-protein predictor serving as an early baseline.
- Graph Neural Network (GNN) models:
- GCN and GAT architectures operating on spatial graphs to propagate neighborhood information.
- Multimodal GNN (RNA + H&E): Node features combine visual embeddings and RNA embeddings to jointly model morphological and molecular structure.
- Adapter-enhanced GNN: A lightweight Adapter Fusion module improves cross-modality integration and stabilizes training across variable tissue regions.
- Patch-based sampling: Mini-batching over subgraphs improves GPU efficiency and alleviates noise in highly heterogeneous slides.
Results
Our best multimodal GNN model achieves state-of-the-art performance on the STP Challenge leaderboard, reaching a Spearman correlation of ~0.74, ranking 1st on the public leaderboard at the time of submission. You can view the result page.
Key findings include:
- Incorporating spatial topology via GNNs substantially outperforms pixel-wise CNN baselines.
- H&E features provide complementary morphological context and improve prediction of proteins with spatial gradients.
- Adapter modules improve robustness and cross-patient generalization.
Impact
This project demonstrates the potential of graph-based multimodal learning for biological tissue modeling, and provides a reproducible pipeline for future research in AI for Science, spatial omics, and cross-modal biological inference.